
 |
The downloadable report file (PDF or text file) for a completed HaloPlex design contains summary information on the targets and amplicons. The downloadable BED-formatted track files contain specific genomic regions to help you analyze the design and compare it to your target regions. This help topic describes the terms used in the PDF report and relates them to the regions in the BED files.

Figure A: Summary information for targets and amplicons as displayed in a HaloPlex PDF report
Figure A is from the PDF report of an example HaloPlex design. The terms in green are defined in the following table.
Target Summary |
|
Region Size |
The Region Size is the total size (in kbp) of all target regions after merging any overlapping regions. The BED file [Design ID]_Regions.bed contains all the merged target regions. The Region Size is also provided in the design details window as the size listed under Target Regions. |
Amplicon Summary |
|
Total Amplicons |
This number is the total number of PCR amplicons expected for the design. The BED file [Design ID]_Amplicons.bed contains the regions of all the expected amplicons. See Figure B for a schematic. The file includes one amplicon per probe. (Note that this differs from the gff files that were previously provided in the original Halo Genomics design wizard. Those files did not include wobble probes.) The Total Amplicons number is also provided in the design details window next to # Amplicons. |
Total Target Bases Analyzable |
The Total Target Bases Analyzable is the number of kilobases in the target regions that are capable of being sequenced and analyzed. To locate these regions, SureDesign identifies the intersection between the covered regions (described further under Total Sequenceable Design Size) and the target regions. Note that the bases in the Total Target Bases Analyzable are sometimes referred to as covered bases. In the HaloPlex design report shown in Figure A, the Total Target Bases Analyzable is slightly smaller than the Region Size, indicating that the sequencing will have some missed target regions. See Figure B for a schematic. The covered regions are those in the [Design ID]_Covered.bed file. The target regions are those in the [Design ID]_Regions.bed file. The Missed track in the [Design ID]_AllTracks.bed file contains the missed target regions (i.e. target regions that are not included in covered regions). |
Total Sequenceable Design Size |
The Total Sequenceable Design Size is the size of the genomic footprint for all sequenceable regions. SureDesign calculates this size by extracting the platform-specific read length from the 5' and 3' ends of each amplicon (or 3' end only for Ion Torrent), and then merging the overlapping regions. For longer amplicons, a portion of the amplicon may not be sequenceable. For example, a 400-bp amplicon sequenced on a MiSeq instrument with 150 PE sequencing would have two sequenceable regions of 150 bp separated by a 100-bp gap. In Figure B, the lighter blue region within the long amplicon represents such a gap. In the HaloPlex design report shown in Figure A, the Total Sequenceable Design Size is larger than the Region Size because the amplicons extend beyond the boundaries of the target regions. See Figure B for a schematic of how target regions, amplicons, and sequenceable regions relate to each other. The BED file [Design ID]_Covered.bed contains the covered regions, which are the sequenceable regions with 5 bases removed from each end (or from the 3' end only for Ion Torrent). The program removes the first 5 bases of sequencing because those bases are more likely to be subject to reference bias when detecting variants. The Total Sequenceable Design Size is also provided in the design details window as the size listed under Sequenced Regions. |
Target Coverage |
The Target Coverage is the percentage of bases in the target regions that are included in the Total Target Bases Analyzable. The covered regions are those in the [Design ID]_Covered.bed file. The target regions are those in the [Design ID]_Regions.bed file. The Target Coverage percentage is also provided in the design details window next to Coverage under Sequenced Regions. |
Recommended Minimum Sequencing per Sample |
This value is the recommended minimum sequencing depth for the HaloPlex design. SureDesign calculates this value by multiplying the Total Sequenceable Design Size by 200. This depth is normally sufficient to cover 90% or more of the target region with 20 reads or more. The exact amount of sequencing needed can only be determined empirically, and it may be higher or lower than the Minimum Sequencing value in the report depending on the genomic region that is amplified. |
The figure below shows a schematic of a genome browser to demonstrate how the regions described in the report files and in the BED files relate to each other.
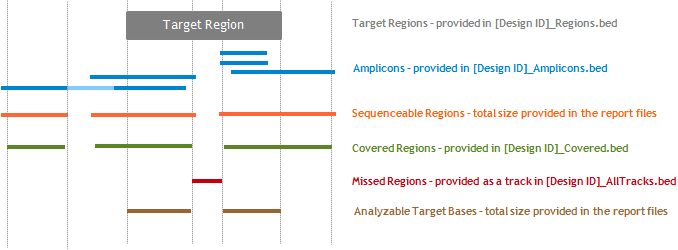
Figure B: Schematic of a genome browser showing a target region, the amplicons from a HaloPlex design to sequence the target region, and other regions referred to the design report files and BED files.
See Also