
 |
 The functionality described in this
topic is only available when you mark Show
Advanced Options.
The functionality described in this
topic is only available when you mark Show
Advanced Options.
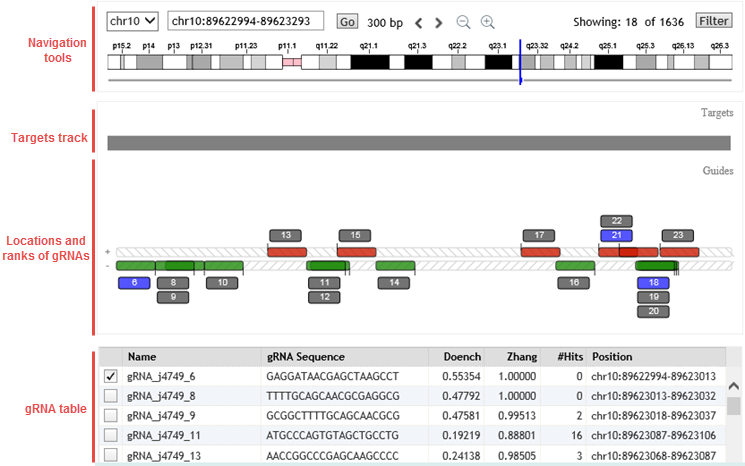
This step includes a genome browser, like the one below, that depicts the locations of the selected gRNAs within a contiguous target region.
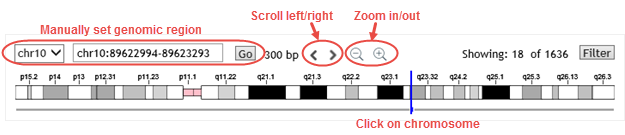
Navigation tools
Use these tools to change the genomic location displayed in the browser.
Filter tool
Click Filter to open the Configure Filters dialog box where you can filter the selection of gRNAs based on the Zhang score.
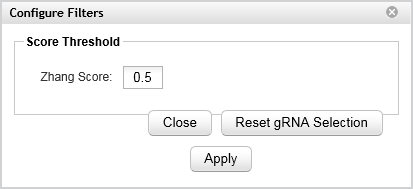
In the Zhang Score field, type the minimum Zhang score for the gRNAs that you want to select, then click Apply. SureDesign de-selects any currently-selected gRNAs that do not meet the specified minimum. Note that lowering the Zhang Score below the default value does not cause SureDesign to select additional gRNAs.
To set the selected gRNAs back to their original settings, click Reset gRNA Selection.
Locations and ranks of gRNAs
The red and green boxes indicate the locations of the gRNAs. The red boxes are gRNAs on the (+) strand of the target, and the green boxes are gRNAs on the (–) strand of the target. Because gRNAs may overlap each other, the boxes are semi-transparent to allow for showing overlapping regions.
For each gRNA shown, there is a blue or gray box that displays the rank number of that gRNA. Blue boxes are used for gRNAs that are currently marked in the table. Gray boxes are for gRNAs that are not marked. Click here for information on how SureDesign ranks gRNAs.
When gRNAs overlap, the boxes displaying the ranks are stacked. For gRNAs on the (+) strand, the bottom number in the stack indicates the left-most gRNA. For gRNAs on the (–) strand, the top number in the stack indicates the
Click directly on a red or green box or a ranking number to select the row for that gRNA in the table at the bottom of the wizard window.
gRNAs table
The table at the bottom of the window lists the following information about each gRNA that SureDesign was able to locate for the target. For those gRNAs that you want to include in the final design, mark the check box in column 1.
Column |
Description |
Check
box |
Mark the check box to include the gRNA in the final design. Any gRNAs not marked in this column are not included in the final design, however, they are still listed in the AllgRNA text file that is available for download after finalizing the design. See SureGuide gRNA design files available for download. |
Name |
The Agilent-assigned name for the gRNA. The naming convention is gRNA#N, where N is a number based on the distance of the gRNA from the start of the target window. |
gRNA Sequence |
The species of the target DNA. The genome build is indicated in parentheses. |
Doench |
The on-target Doench score calculated for the gRNA. |
Zhang |
The off-target Zhang score calculated for the gRNA. |
#Hits |
The number of sequence matches for the gRNA within the genomic region currently displayed in the browser. |
Position |
The genomic coordinates of the gRNA. |
After you review the gRNAs, mark the ones that you want to include in the final design. Then, click Back to return to the Summary step and click the Review link for the next region in the list.