The basic elements of a microarray are , and eArray
provides many ways for you to define the probes that you want. One of
the main ways is to search for them. In eArray, you can retrieve probes
of interest with a variety of search methods. In addition, a probe search
provides an easy way to construct a probe
group based upon your own criteria. You then use one or more probe
groups to create a microarray design. See Why
create probe groups.
You can customize eArray to pre-set certain search parameters. You can
also customize the content and column order of search results. See Set user preferences.
This help topic contains the following sections:
eArray probe
search tools
Choosing
a search tool
Additional
guidance from Agilent
eArray probe search tools
You can search the probes in the eArray probe database in several ways:
Choosing a search tool
For probes that are already designed, the
search methodology that you use to find probes depends upon your level
of familiarity with the target sequences, and the specific kind of study
that you want to do. For example, if you want to do a differential gene
expression study, but you do not know specific transcript or gene identifiers,
you can use GO Search to return probes
for genes based upon biological process or molecular function.
You could also use Search
Standard Probes to take a similar, but less structured approach. When
you enter one or more keywords as search criteria, you can search probe
annotation, which often describes a target gene's function. A search can
return all probes whose annotation contains the word or phrase you typed.
If you know the specific targets for which you want probes, you can also
use Search Standard Probes to advantage. You can upload lists of GenBank
IDs or gene symbols, and return all probes that
match the criteria. This allows you to be very specific about your search
criteria.
Each probe search tool has distinct advantages,
as described in the table below.
Search
tool |
Comments |
Search
Standard Probes |
This
tool gives you many options to retrieve the probes that you want.
You can type a single search term, and the search can return all
probes that contain the term in any of their annotation. This
is a good search methodology to use when you first explore the
content of the database, and want to see what types of probes
exist in the system. For example, you can type the term kinase,
and the search returns all probes that have this word in their
annotation, including probes with annotation such as protein
kinase C, delta,
and hexokinase 3.
You can also select a specific type of annotation or accession,
and enter one or many search terms of that type. This is most
useful when you want to use identifiers such as probe IDs, GenBank IDs, or gene symbols. You can simply
upload a list, and the search returns all probes that match. |
GO
Search |
A
Gene Ontology (GO) Search is a good way to identify probes for
genes and gene products that are associated with biological processes,
molecular functions, and/or cellular components that you want
to investigate. You enter a standard GO term, and the search retrieves
all of the probes that are associated with the term. eArray can
also help you find relevant GO terms to use in the search. This
type of search is available for standard Expression probes. |
HD
Probe Search |
You
can search the HD CGH and ChIP databases to return probes within
specified genomic regions, at a very high density. You can then
use these probes to create a microarray that has higher resolution
than is seen for catalog array offerings for these applications.
To do an HD search, you define the desired genomic regions, and
the density or number of probes that you want. |
SNP
Probe Search |
This
specialized search is the only way to retrieve Agilent SNP probes,
which are probes that are designed specifically for Agilent CGH+SNP
microarrays. You can use CGH+SNP microarrays to deduce the genotypes
of SNP sites, calculate allele-specific SNP copy numbers, and
find regions of loss of heterozygosity (LOH). This search is available
in the CGH application type. For more information, see CGH+SNP
Microarrays. |
Exon
Probe Search |
This
specialized search is the only way to retrieve Agilent Exon probes,
which are probes that are designed specifically for Gene Expression
Exon microarrays. You can use these microarrays to study differential
splicing between tissues, between disease and non-disease states
(i.e. cancerous vs. non-cancerous), and between different forms of
the same disease. This search is available in the Expression application
type. For more information, see Exon
microarrays. |
Additional guidance from Agilent
How do
I create a complex CGH search that yields probes at different resolutions
for different genomic regions and also with different filtering criteria
for different regions?
For the
CGH application, what are the advantages and disadvantages of using the
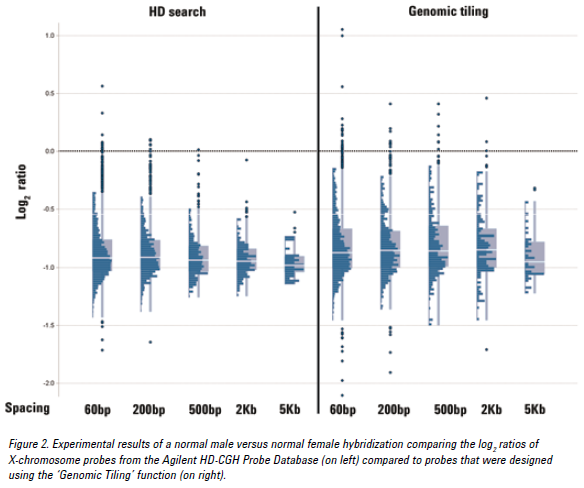
‘Genomic Tiling’ function (under the “Probe” tab) compared to using Agilent
HD-CGH Database Probes?
When
designing CGH microarrays, how can I avoid GC-rich, high-Tm
or repeat regions?
In the
CGH HD Probe Search, which ‘Similarity Filter’ will work for my design?
What are the consequences of using or not using the filters?
In the CGH HD Probe Search, how
do I target exons only, not just genes?
See also
Overview
of working with probes in eArray