Overall, probes generated using the ‘Genomic Tiling’ function will perform more poorly than probes found in the Agilent HD-CGH Probe Database. Agilent very strongly recommends using the Agilent HD-CGH Probe Database and not the ‘Genomic Tiling’ option. Only for regions where there are not enough HD probes available in the database should ‘Genomic Tiling’ be considered.
All HD probes in the database (except for probes in regions in which no optimal Tm probes exist) have been Tm matched and have a predicted performance score (based on Tm, GC content, a hairpin ΔG, sequence complexity, and metrics to measure homology with the rest of the reference genome). The eArray pair-wise reduction algorithm will pick the best HD probes based on the user-selected average HD probe spacing per interval or the total number of HD probes.
Additionally, during design the HD probes have passed a Tm filter, are annotated such that a user can choose between different similarity filtering options (non-unique probe filter, perfect match filter, or similarity score filter), and if there are catalog probes present in search results they can be preferentially selected. In contrast, using eArray’s ‘Genomic Tiling’ feature probes are picked at a fixed spacing and there is no chance to Tm balance or optimize probes selected for by performance. Probes created should perform no better than those picked at random. The only options to improve probe performance are probe trimming and skipping of repeat masked regions.
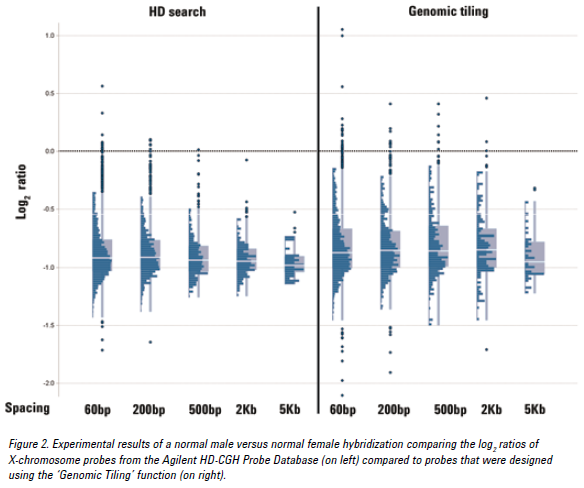
See the figure below for an example of CGH data from Agilent HD-CGH probes compared to ‘Genomic Tiling’ probes. The median log2 ratios of the HD-CGH probes are closer to the expected value of -1 with a smaller spread when compared to the ‘Genomic Tiling’ probes.