Learn about mutagenic oligos and mutagenesis libraries |
|
In eArray, mutagenesis libraries are collections of DNA oligonucleotides (oligos) that can be used to create families of variant genes. These genes can then be used to produce families of variant proteins for functional studies. Traditional mutagenesis methods that use radiation or chemical agents can only produce random mutagenesis, and the reactions are difficult to control. Standard oligonucleotide-directed site-specific mutagenesis methods can produce well-defined sequence changes, but the creation of large numbers of mutants can be both time- and cost-prohibitive. In contrast, the mutagenesis oligo libraries that can be created in eArray give you precise, codon-level control over the variants that are produced. Use the oligo libraries together with the Agilent QuikChange Oligo Library Mutagenesis System reagent kit to quickly create large libraries of well-defined mutants that are ideally suited for applications such as scanning mutagenesis and mutagenesis that is targeted to many specific amino acids of interest.
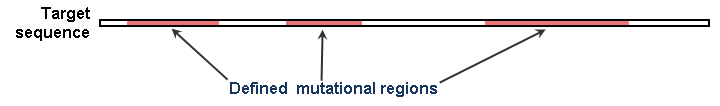
eArray designs mutagenic oligos to target sequences that you enter. Target sequences can be genes or other DNA sequences of interest. Within each target sequence, you define non-overlapping mutational regions to which to apply one or more mutational strategies. See the figure below for an example.

Figure 1 – Target sequence with mutational regions defined. You can apply one or more mutational strategies to each mutational region.
Mutational strategies create families of sequences that alter codons in the target sequence. You apply mutational strategies on the amino acid level. Click here for descriptions of the mutational strategies available in eArray.
When you set up a mutational strategy, you also select the specific type of protein expression host that you intend to use. This lets you customize the codon usage for the mutated positions in the generated mutagenic sequences to maximize the efficiency of expression. eArray supports E. coli, yeast, mammalian, and insect host types. By default, eArray uses the most commonly-used codons in the selected protein expression host for each mutated amino acid. Alternatively, you can customize the codon usage for individual amino acids.
To the mutant sequences that are generated for a given mutational region, eArray adds 5' and 3' sequences that include PCR primer annealing sites. The annealing sites accommodate custom PCR primers that are used later in the mutagenesis protocol to amplify the mutagenic oligos that are generated for the given mutational region. eArray optimizes both the length and position of the PCR primer annealing sites based on several criteria. You can order the PCR primers that eArray designs for your library from an oligo manufacturing vendor.
For each mutational region, the result is a set of homologous mutagenic oligos that have identical 5' and 3' end sequences. In all cases, the sequence of a mutagenic oligo exactly matches the corresponding region of the target sequence, except at the location(s) of the requested change(s). The mutagenic oligo set created for a representative mutational region appears in Figure 2, below.
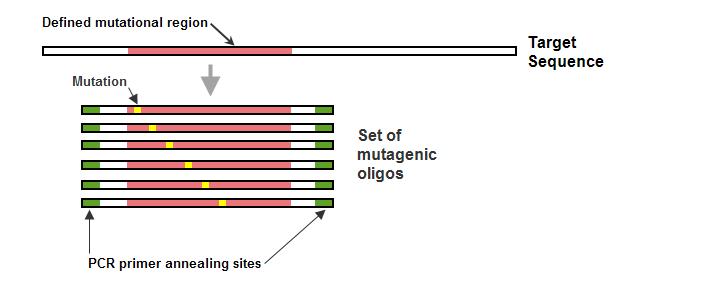
Figure 2 – Set of mutagenic oligos (oligo set) generated for one mutational region in a target DNA sequence. All oligos in the set are the same length, and are identical in sequence to the corresponding region of the target sequence, except for the necessary base changes to create the requested amino acid changes. The mutational regions in the oligos are flanked on both the 5' and 3' ends with sequences that contain PCR primer annealing sites. If space on the oligos and within the target sequence allows, eArray can optimize both the length and position of the PCR primer annealing sites for the set.
Target sequence – A target sequence can be from 110 to 50,000 bp in length. Within this sequence, the 30 nucleotides on the 5' and 3' ends of the sequence cannot be included in a mutational region. These flanking sequences accommodate PCR primer annealing sites. However, it can be advantageous to enter target sequences with longer flanking sequences, since this can give eArray greater flexibility to optimize both the length and position of the PCR primers.
Length
of mutational regions –
Minimum: 17 amino acids
Maximum: The cumulative total for all mutational regions in the library
is 1000 amino acids. If you define a mutational region that is longer
than 50 amino acids, eArray divides the defined region into subregions
of roughly equal size (no larger than 50 amino acids each). Each subregion
generates its own pair of PCR primers and its own mutagenic oligo
set. The effective size range of target sequence covered by a single
mutagenic oligo set is 17 to 50 amino acids.
Number of defined mutational regions – Up to 20 non-overlapping mutational regions of 17 to 50 amino acids each for a given target sequence.
Length of PCR primers – 18–28 nucleotides
Length of mutagenic oligos – 100–200 nucleotides (51–150 nucleotides of which represent the given defined mutational region.)
Maximum number of oligos per library – Currently, up to 27,500
Control oligos included in libraries – Agilent also automatically includes LacZ control oligos in every custom mutagenesis library. You use these oligos, in conjunction with matching PCR primers that are provided with the kit, as positive controls to assess library titer and mutational efficiency. They are also used by Agilent for quality control purposes.
eArray groups all of the oligos that are created from a given mutational region with any mutational strategy into an oligo set. Oligo sets are collections of homologous sequences that have the same 5' and 3' ends that serve as PCR primer annealing sites. A mutagenesis library can contain 1 to 20 oligo sets.
The figure below illustrates an example of a mutagenesis library structure.
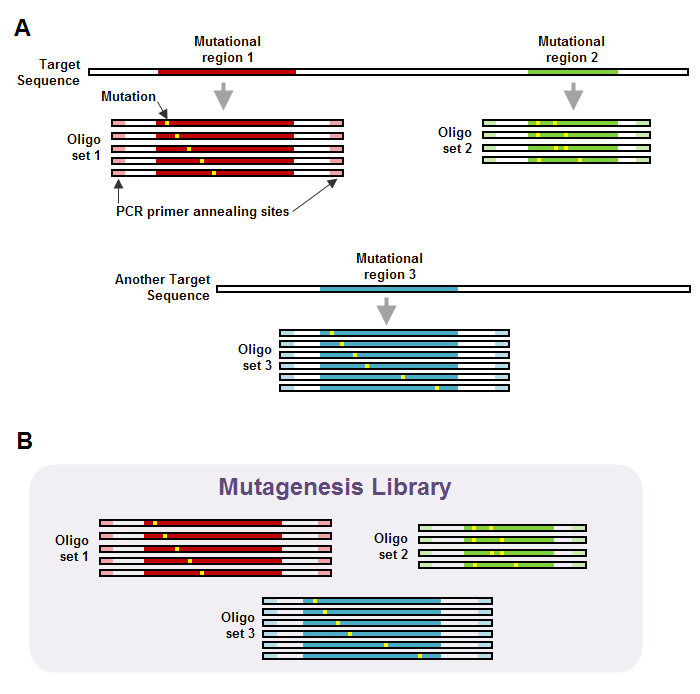
Figure 3 – Structure of a mutagenesis library. A – Two target sequences. Two mutational regions have been defined for one target sequence, and one mutational region has been defined for another. This generates a total of three oligo sets. Each oligo set contains a separate oligo for each variant of the mutational region that is produced by the selected mutational strateg(ies). Note the PCR primer annealing sites on the 5' and 3' ends of the oligos. B – Mutagenesis library created from the two target sequences in panel A. The mutagenesis library contains all of the oligo sets that were generated from the target sequences. The PCR primers that eArray designs for the library need to be purchased through an oligo manufacturing vendor.
To create a mutagenesis library, see Ways to create mutagenesis libraries.
To learn how to navigate the Mutagenesis application type, see Learn to navigate the Mutagenesis application type.
Each mutagenesis library has a status that determines the actions that you can take on it. You can select one of these statuses when you create a library:
Draft – The library is in its initial stages of development. As the owner of the library, you can make and save changes to it. If desired, you can submit the library to Agilent Manufacturing, which changes the status of the library to Submitted.
Submitted – The library is in its finished form, and has been submitted to Agilent manufacturing. You cannot further edit the library, or change its status to Draft.
Note: When you submit a mutagenesis library to manufacturing, an Agilent representative contacts you to follow up on your interest. Submitting a library does not obligate you to place an order for the library.
The table below shows the specific actions that you can take on mutagenesis libraries of each status. Click the links for more information on each action.
Action |
Draft |
Submitted |
● |
● |
|
● |
● |
|
● |
|
|
● |
* |
|
|
● |
|
|
● |
|
● |
|
* Libraries with Submitted status have already been submitted to Agilent Manufacturing.
See also