
 |
The CustomFISH wizard allows you to create FISH probes to detect a single target region of interest in the human or mouse genome. The wizard asks you to define the target region using a gene name or other identifier and then walks you through the selection of parameters and options. You then submit the design to SureDesign and the program's algorithms select the oligo sequences based on the information you provided. When your oligo selection job is complete, you receive an e-mail notifying you that your design is ready to be finalized. Once you finalize the design, it is available for ordering.
To open the CustomFISH design wizard:
· At the top of the screen, click Create Designs > CustomFISH.
The wizard window opens to Step 1.
The steps of the CustomFISH wizard for a single probe design are described in detail in this help topic. The steps are:
Step 7: Finalize/Design Complete
In this step, complete the fields described below to define the design.
Design Name
Type a name for your design into the field. Alphanumeric characters, hyphens, underscores, and spaces are permitted. The name must be no more than 30 characters and it must be unique within your workgroup.
Species
Specify the species of the design. The default selection is H. sapiens, but CustomFISH designs can also be created for M. musculus.
To change the selection, click Select. The Select Species dialog box opens, allowing you to select the desired species.
Build
If multiple genome builds are available for the selected species, select the desired genome build in the provided drop-down list.
If no drop-down list for build selection is provided, that indicates that only one genome build is currently available for the selected species. That build is indicated below the Species field.
Specify the folder in which you want to save this design. The default selection is the top-level folder for your workgroup.
To change the selection, click Select to open the Select Folder dialog box, and mark the folder in which you want to save the new design. This dialog box lists the available folders within your workgroup and, if you are a member of any collaborations, lists the collaboration folders to which you have access.
Category
Select Single Probe. This category is for the creation of designs for a single target of interest.
NOTE For instructions on creating a break apart or dual fusion design, see Create a CustomFISH break apart design or Create a CustomFISH dual fusion design.
Design Type
Specify if you want the design to be shared or private.
· Select Shared to allow SureDesign to make the oligos in your design available to other SureDesign users. Selecting Shared does not mean that Agilent makes your design, your design information, or any of your personal information available to other customers.
· Select Private to restrict SureDesign from recommending your exact design to other users. Note that private designs could have a longer lead time as they will be prioritized below shared designs. Agilent reserves the right to sell oligos with overlapping or matching coordinates to other customers, regardless of shared or private design status.
Click Next to advance to Step 2.
In this step, define the target region that you want to detect with the CustomFISH design.
NOTE You can only define one target region with the CustomFISH wizard. Run the wizard again to create designs for additional targets of interest.
In the Targets text area, type or paste the target identifier directly into the text area. For single probe designs, you can enter the target identifier using one of the three identifier types described in the table below.
Table: Permitted identifiers for defining targets
Identifier type |
Description |
Examples |
Target gene |
A single NCBI gene symbol, gene ID, or accession number. |
|
Genomic coordinates |
The genomic coordinates (UCSC browser format or BED format) for a single genomic region. |
|
Cytoband |
A cytoband identifier. |
|
Click Next to
advance to Step 3.
In the Enter Parameters step, you can set the size of the left and right padding regions. Because the size of the design input region needs to be between 100 and 2000 kb, padding is required for targets <100 kb in order to increase the size of the design input region. SureDesign includes the padding regions in the oligo selection job.
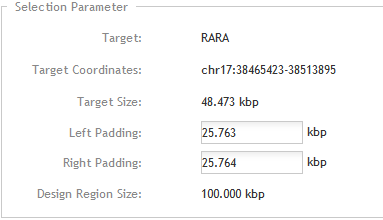
Target
This field displays the target as you entered it in the previous step.
Target Coordinates
This field displays the genomic coordinates of the target region.
Target Size
This field displays the size of the target region. If the target region is less than 100 kb, padding is required to increase the total design size.
Left Padding
Enter the size of the padding region to the left of the target.
Right Padding
Enter the size of the padding region to the right of the target.
Design Region Size
This field displays the size of the total design input region, which includes the target region plus the left and right padding regions. If you change the value in the Left Padding or Right Padding field, the Design Region Size field updates automatically. The size must be 100-2000 kb.
Click Next to
advance to Step 4.
At this step, review the Target Details before you proceed to submit the design for oligo selection.
The Target Details table lists the following information for the design input region.
· Target ID - The Target ID column lists the target identifier as entered in the Define Targets step.
· Base Pairs - The Base Pairs column lists the total number of base pairs in the design input region.
· Position - The Position column displays the genomic coordinates for the design input region (including any padding) as link. Click the link to view the region in the UCSC Genome Browser.
Click View targets in UCSC to open the UCSC Genome Browser and see the genomic location of the design input region.
Click Next to advance to Step 5.
At this step, SureDesign searches its database of catalog and shared CustomFISH designs to check for any existing designs that it can recommend to cover your design input region. You then have the option of selecting one of the recommended designs or continuing to create your own. Because the recommended designs are already available in Agilent manufacturing, you can receive your design faster by selecting a recommended design.
Your Design
The Your Design table at the top of the wizard window shows the design name and the design input region, which includes your target region and any padding.
Recommended Designs
The Recommended Designs table at the bottom of the wizard window lists the existing designs recommended by SureDesign that cover at least 70% of your design input region. The information provided in the table is described below.
· Design ID - The unique ID number for the recommended design (shared customFISH designs only)
· Catalog ID - The unique ID number for the recommended design (catalog FISH designs only)
· Design Region - The genomic coordinates of the region covered by the recommended design
· Design Size - The total size of the design
· Overlap - The percentage of overlap between the design input region and the recommended design
· Action - Contains a link for downloading the design files
To select a design:
Review the recommended designs to decide if any meet your needs. To help you decide, you can download the design files for a recommended design (click Download in the Action column) or view the overlap of the recommended designs with your design input region in the UCSC Genome Browser (click View Design Overlap in UCSC Browser).
Mark the check box next to the design that you want to use.
· If you want to use one of the recommended designs, mark the check box next to the design.
· If you want to continue creating your own design, mark the check box in the Your Design table.
To submit the design for oligo selection:
After you select a design, submit the design to the SureDesign job queue and the SureDesign algorithms will select the oligos for your design.
Click Begin
Oligo Selection.
A message box opens indicating the e-mail address where Agilent will
contact you when the oligo selection job is complete. If desired,
you can enter additional e-mail addresses into the provided field.
Click OK
in the notification message to submit the design to SureDesign.
Your submission is placed in the SureDesign job queue to await oligo
selection.
The wizard automatically advances to Step 6.
At this point in the design creation process, SureDesign is processing your oligo selection job. The length of time required for SureDesign to complete the job depends on the number of jobs waiting in the queue and the size of your design.
Click Close Design Wizard. When you receive an e-mail from Agilent SureDesign notifying you that your oligo selection job was successfully completed, relaunch the wizard and continue creating the design:
Open the SureDesign Home screen.
Locate the design under Designs:
In Progress, and click the Continue icon ![]() .
.
The wizard window opens to Step 7.
NOTE You can monitor the status of your oligo selection jobs from the SureDesign Home screen.
In this step, click Finalize Design to finish the design creation process. Alternatively, if you want to make further edits to the design, click Modify Design to delete the oligos from the oligo selection job and return to the Define Targets step.
At this step, the information in the design summary panel is complete, and you can download and view the summary files to help you decide if you want to finalize the design or revise it further.
After clicking Finalize Design, the wizard window updates to the Design Complete step, and provides the following information:
Design Name |
The name of the design. |
Design ID |
The unique ID number for the customFISH design. |
Catalog ID |
The unique ID number for the catalog FISH design. |
Design Region Size |
The total size of all genomic regions covered by the design. |
Coverage |
The percentage of nucleotides in each probe design that are covered by one or more oligos. The covered region is included as a track in the Covered BED file for the design. |
Use the action buttons at the bottom of the Finalize Design window to take
further action on the design:
· Click Order to open the Order dialog box, where you can add the design to your Agilent Genomics shopping cart or submit a request to receive a price quote.
· Click Mark as Favorite to add the design to your list of favorites. The design will appear in the Designs: Recent and Favorites dashboard on the Home screen.
· Click Download to download one or more design files, including a formatted PDF report that summarizes key information on the design, oligos, targets, and the oligo selection job.
These action buttons are also available from the design details window.